The human immune response and the impact of age, sex, and genetics
Welcome! This work is brought to you by the Milieu Interieur Consortium and is published by Piasecka and Duffy et al., 2017.
ABSTRACT Immune responses are highly variable between individuals and populations, with this variance mediated by many factors including age, sex, and genetics. To dissect how each of these factors contributes to differential immune responses we recruited 1,000 healthy donors as part of the Milieu Intereiur cohort that were equally stratified by age (20-70 years old) and sex (50:50). Whole blood from each donor was stimulated in a standardized approach with Escherichia coli, BCG, Staphylococcus aureus, SEB, Candida albicans and Influenza virus and the transcriptomic response was analyzed by Nanostring gene expression arrays. We show that age affects gene expression in a stimuli specific manner, while the effect of sex is more common across conditions. Both age and sex contribute to the expression of many immune-related genes but at a low level, while in contrast, the genetic effects are much stronger but relevant to fewer genes. Genetic analysis identified multiple eQTLs, both local and trans, for all stimuli induced responses, including a novel, strong trans-eQTL located near the CR1 gene after C. albicans stimulation. Finally, we employed a structural equation model to describe how age and sex effects are mediated by different immune cell populations. These results lay the foundations for understanding how age, sex, and genetics may impact differentially induced immune responses, which may be considered for future clinical strategies proposing stratified or precision medicine approaches.
Study outline
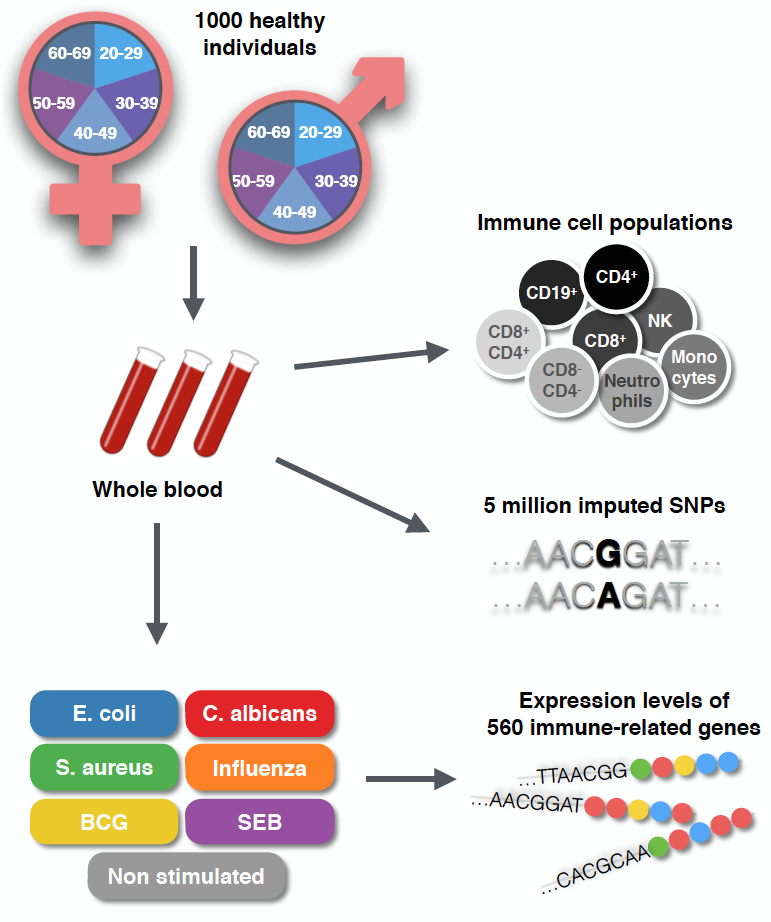
Transcriptomic response
Click-Drag to zoom, Shift-Click-Drag to rotate; Use 'Options' below to adjust subsampling.Specificity of age and sex effects on gene expression
The effect of age is stimulus-specific with ~40% of genes presenting an age-dependent expression pattern in only one or two conditions. Conversely, sex effects on gene expression are more frequently shared across stimuli with ~45% of genes differing in their expression between sexes in at least six or seven conditions.Overview of genotypic effects on gene expression
135 Genes were found with local Cis eQTLs in the unstimulated state and around 140 genes presenting an eQTL after stimulation. On the other hand, trans-regulation was noticeably stronger after stimulation, particularly with E. coli, BCG, C. albicans and SEB.Quick gene view
Table search
The human immune response and the impact of age, sex, and genetics
This work is brought to you by the Milieu Interieur Consortium and is published by Piasecka and Duffy et al., 2017. with the following title, authorship, and affiliations:
Impact of age, sex and genetics on transcriptional immune responses to bacterial, viral and fungal challenges. Barbara Piasecka 1,2* , Darragh Duffy 2,3,4* , Alejandra Urrutia 3,4,5 , Helene Quach 1,6,7 , Etienne Patin 1,6,7 , Céline Posseme 2 , Bruno Charbit 2 , Vincent Rouilly 2 , Cameron MacPherson 2 , Milena Hasan 2 , Benoit Albaud 8 , David Gentien 8 , Jacques Fellay 9,10 , Matthew L. Albert 2,3,4,5,# , Lluis Quintana-Murci 1,6,7,# , for The Milieu Intérieur Consortium † .
- 1 - Laboratory of Human Evolutionary Genetics, Department of Genomes & Genetics, Institut Pasteur, Paris, 75015, France
- 2 - Center for Translational Research, Institut Pasteur, Paris, 75015, France
- 3 - Laboratory of Dendritic Cell Immunobiology, Department of Immunology, Institut Pasteur, Paris, 75015, France
- 4 - INSERM U1223, Paris, 75015, France
- 5 - Department of Cancer Immunology, Genentech Inc, San Francisco, CA 94080, USA
- 6 - CNRS URA3012, Paris, 75015, France
- 7 - Center of Bioinformatics, Biostatistics and Integrative Biology, Institut Pasteur, Paris 75015, France
- 8 - Institut Curie, Centre de Recherche, Département de recherche translationnelle, Plateforme de Génomique, Paris, 75005, France
- 9 - School of Life Sciences, École Polytechnique Fédérale de Lausanne, Lausanne 1015, Switzerland
- 10 - Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland
- * - Indicates co-first authors
- # - Indicates co-senior authors
- † - The Milieu Intérieur Consortium ¶ is composed of the following team leaders: Laurent Abel (Hôpital Necker), Andres Alcover, Kalla Astrom (Lund University), Philippe Bousso, Pierre Bruhns, Ana Cumano, Caroline Demangel, Ludovic Deriano, James Di Santo, Françoise Dromer, Gérard Eberl, Jost Enninga, Jacques Fellay (EPFL, Lausanne) Antonio Freitas, Odile Gelpi, Ivo Gomperts-Boneca, Serge Hercberg (Université Paris 13), Olivier Lantz (Institut Curie), Claude Leclerc, Hugo Mouquet, Sandra Pellegrini, Stanislas Pol (Hôpital Côchin), Lars Rogge, Anavaj Sakuntabhai, Olivier Schwartz, Benno Schwikowski, Spencer Shorte, Vassili Soumelis (Institut Curie), Frédéric Tangy, Eric Tartour (Hôpital Européen George Pompidou), Antoine Toubert (Hôpital Saint-Louis), Marie-Noëlle Ungeheuer, Lluis Quintana-Murci § , Matthew L. Albert §
- ¶ - unless otherwise indicated, partners are located at Institut Pasteur, Paris
- § - co-coordinators of the Milieu Intérieur Consortium
Data are made available from the electronic version of the publication ( here )
